In a race to train large language models that generate images and engage in dialogue, tech companies spend billions of dollars and millions of household hours in energy consumption [1] every month. Today’s AI systems use neural network architectures such as transformers which often function as ”black boxes” with billions of parameters.
While optimizing these AI systems consumes a significant amount of money and energy, nature has had time to develop more elegant solutions to neural network design. In this sense, let us introduce you to Drosophila melanogaster, the humble fruit fly – whose poppy-seed sized brain demonstrates remarkable computational efficiency that puts any current artificial network to shame.
From navigating complex flight patterns to detecting predators and finding mates, this tiny creature survives seamlessly in the real world. The fruit fly’s brain accomplishes all these parallel computations with unmatched efficiency, using just about 130,000 neurons. It runs these computations on an estimated 120 nanowatts of power – eight times less than the power draw of a quartz watch, which can only track time. This extraordinary efficiency, perfected through millions of years of evolution, enables complex behaviors on a near-zero power budget.
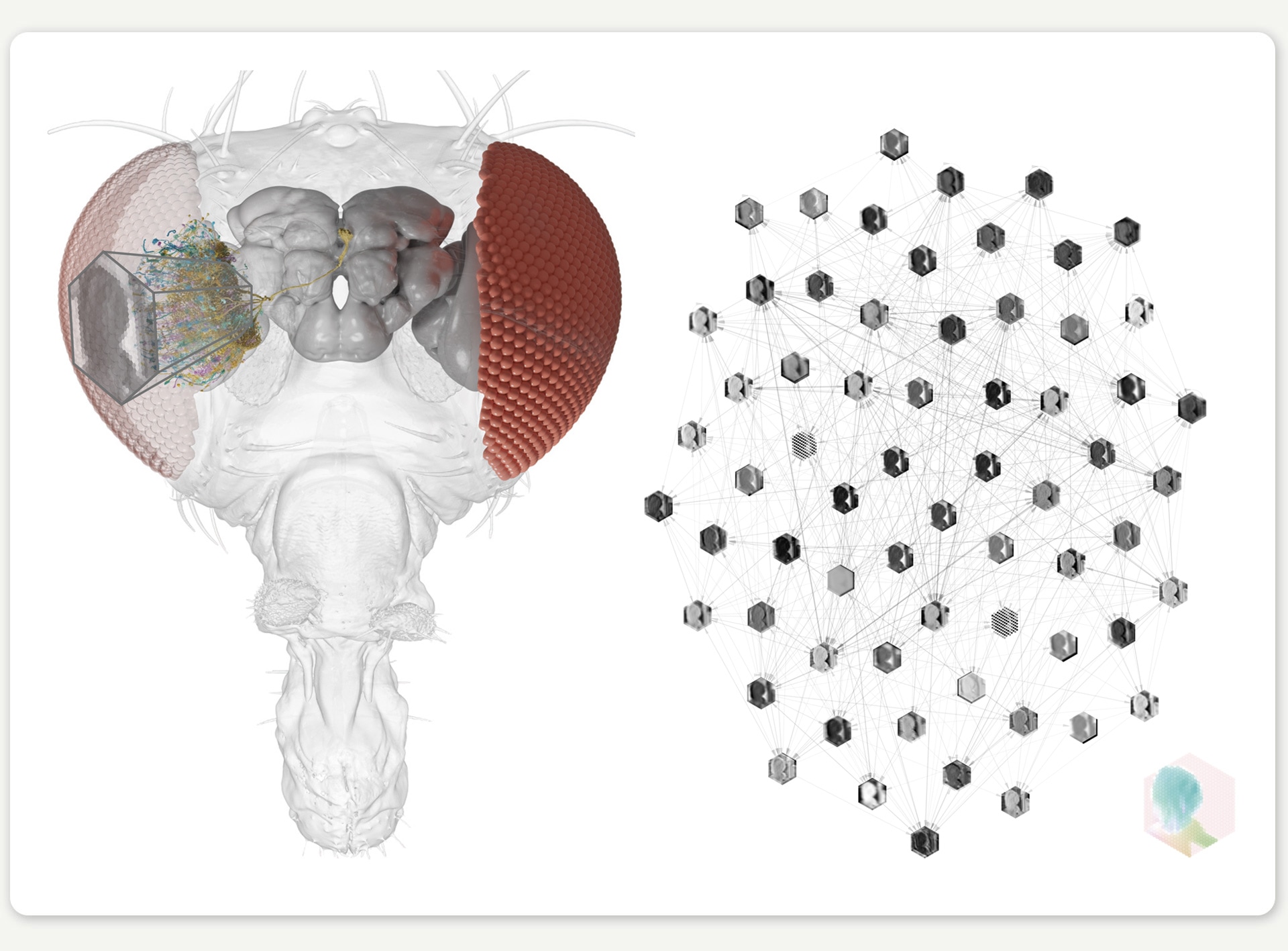
Figure 1: Light enters the compound eye of the fly, causing photoreceptors to send electrical signals through a complex network of neurons, enabling the fly to detect motion. Fruit fly neurons detect motion within 30 milliseconds, initiating escape maneuvers in half the time of a human blink.
Major scientific discoveries around Drosophila
For nearly a century, this insect has been at the forefront of biological discovery. Groundbreaking discoveries have led to several Nobel Prizes in which Drosophila was used as the model system, including the identification of genes controlling circadian rhythms and development. Its rapid reproduction cycle – just 10 days from egg to adult – allows scientists to study multiple generations in weeks rather than years. Genetic tools, like the GAL4-UAS system, enable precise control over phenotypes, including the expression of specific characteristics of neurons. Measurement techniques ranging from electrophysiology to calcium imaging, enable recording neural activity anywhere in the fly. Combined with large-scale behavioral recordings, the properties and tools available for studying the fruit fly make it an ideal testing ground for understanding cause and effect in biological intelligence.
Building on this rich scientific foundation, several research teams (particularly the FlyEM project at Janelia and the FlyWire consortium) have been able to map the fly’s entire connectome to create a comprehensive diagram of every neuron and neural connection in its brain. This digitalized representation of the brain structure provides a complete map for electrical signaling between neurons, and opens up unique opportunities for understanding information processing in a biological neural network.
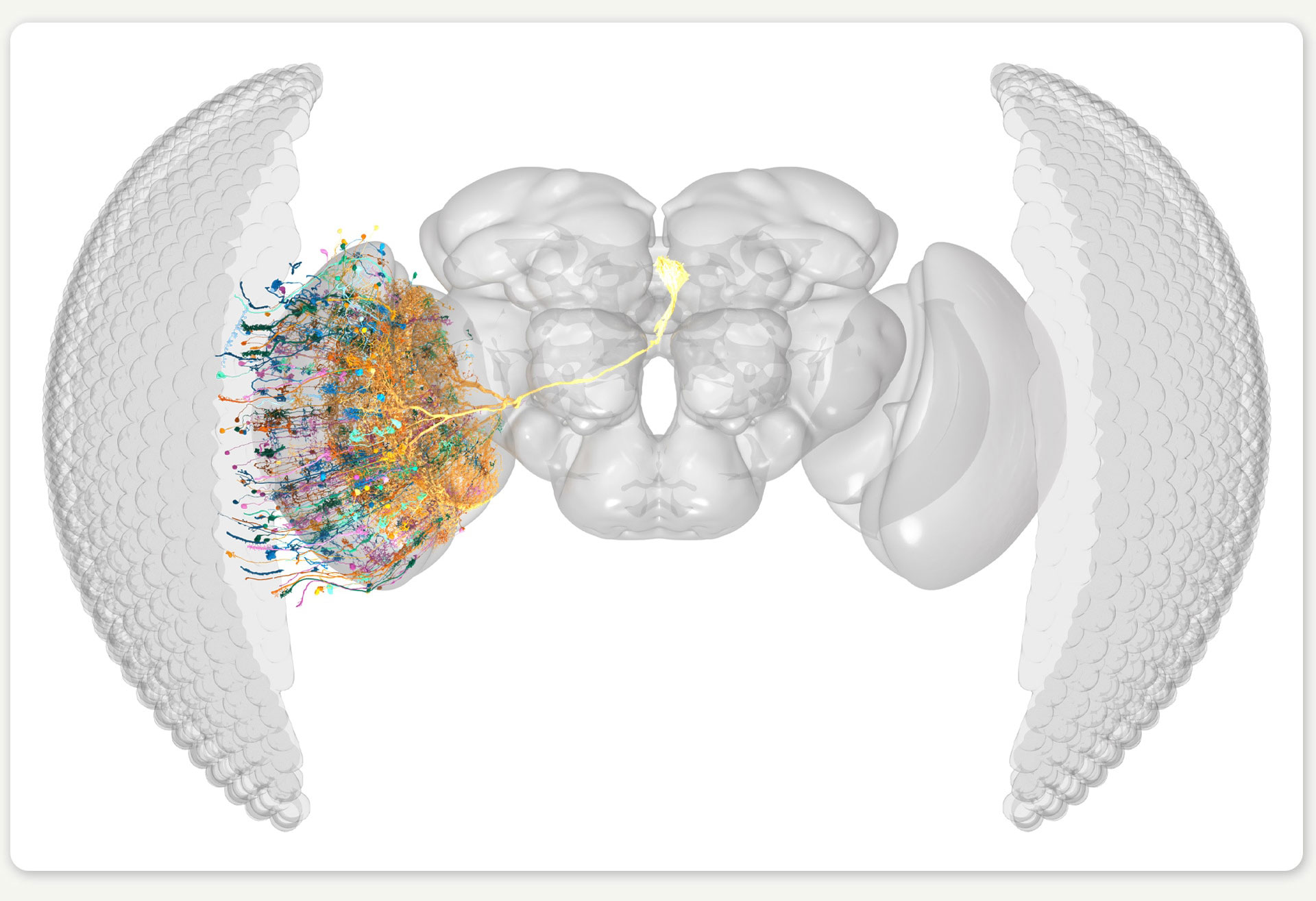
Figure 2: Rendering of the right eye connectome, showing a small number of the neurons for illustration. ©SIWANOWICZ, I. & LOESCHE, F. / HHMI JANELIA RESEARCH CAMPUS
Understanding how a connectome computes
But having a connectome is one thing; understanding how it computes is another entirely. This is where our research, a collaboration between the labs of Jakob Macke (University of Tübingen) and Srinivas Turaga (HHMI’s Janelia Research Campus), comes in. In our work, which was recently published in Nature, we developed a neural network model where each artificial neuron maps to a real neuron in the fruit fly’s brain. In contrast to most artificial neural network models with billions of parameters, our connectome-based model only uses 735 carefully mapped parameters. Given that we did not know the specific parameter values at the start, we began with a guess and then optimized the model to perform a behaviorally relevant task. Because motion detection is important for fruit fly survival, we trained our model on an optic flow computer-vision task to detect motion in naturalistic videos. Through this training, the model’s dynamic computations mimic one of the fly’s core visual abilities.
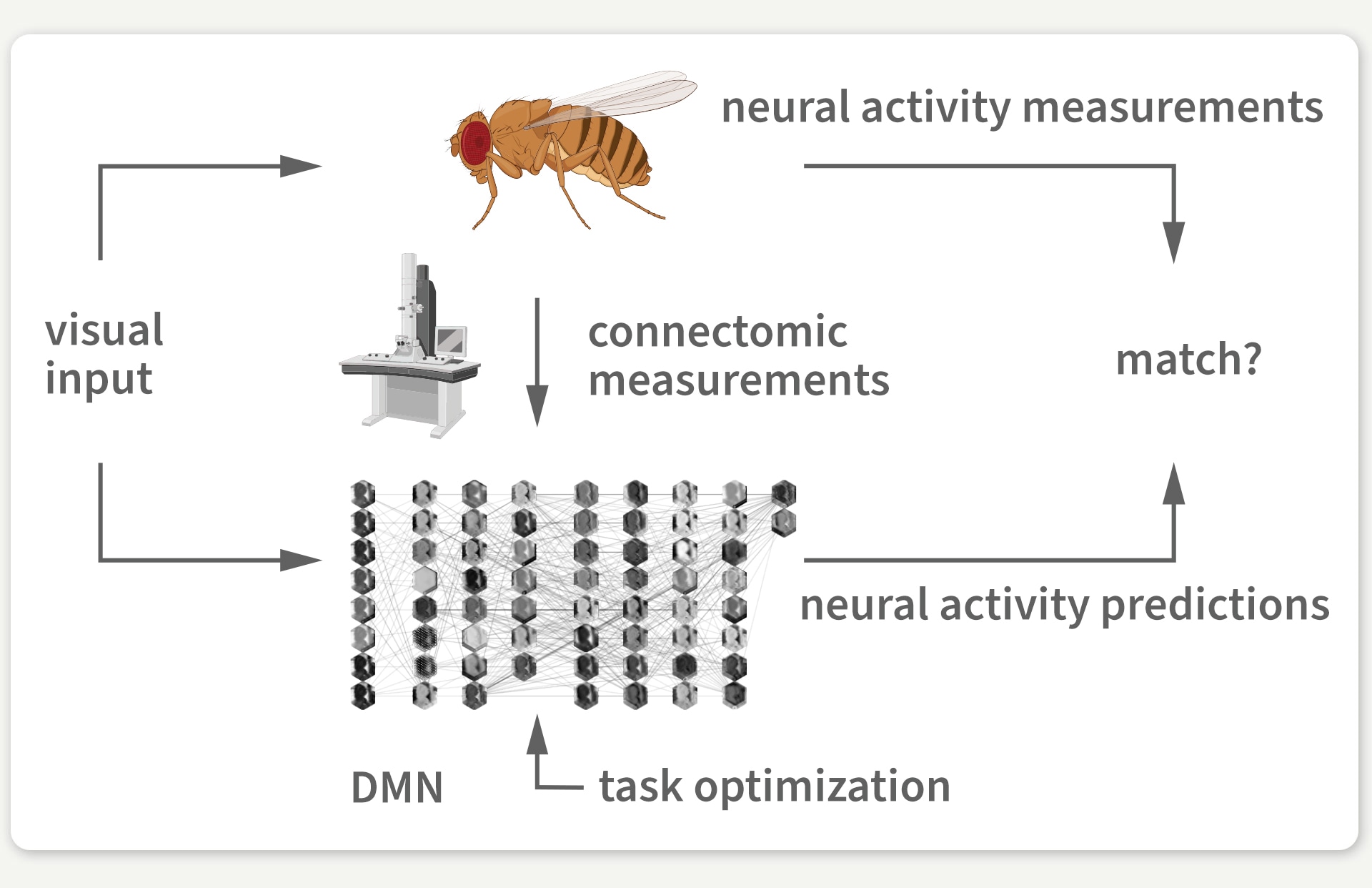
Figure 3: We use the connectome to build a detailed deep mechanistic network simulation of the fly visual system that can predict neural activity in living brains.
Our model is only guided by this one-to-one mapping to real brain neurons, a dynamical neuron model, and the motion detection task. As a result, we had no guarantee that the activity of individual neurons in the model would match actual measurements of neural activity in the animal brain that have been taken in different experimental studies and from different fruit flies. We computationally measured the model’s neural activity in response to laboratory stimuli from real experiments to compare our model’s neural activity predictions with brain measurements at the level of individual neurons.
Luckily, a brain from one fruit fly is strikingly similar to the brain of another fruit fly – and now to our model as well. We found that in response to laboratory stimuli, the activity of each artificial neuron in our model matched the activity of its biological counterpart for which such measurements were available. And we were able to validate our discovery against 26 experimental studies.
Getting here took a lot of work. One of our biggest challenges wasn’t adding complexity – it was learning what we could remove. We started with ambitious goals: modeling multiple visual tasks, implementing complex synapse dynamics, and running numerous comparisons. But the breakthrough came when we carefully reduced our model and addressed the uncertainty that was due to imperfect simulation and optimization. Focusing on essential, evidence-based tasks like optic flow detection, a simple synapse model, and systematic analysis against suitable comparison groups – one computational experiment at a time – allowed us to make the most of the connectome data.
What’s next?
Our model makes predictions for many details of information processing in the fly brain. However, our model still does not simulate many biological details, such as the effects from neuromodulation, glial cells, and electrical synapses. This is just the beginning, and there is a lot about the nervous system that we know how to measure but not how to model yet. Our approach opens up new opportunities for the iterative refinement of models. Over time, we will be able to integrate more measurements, both to gradually advance our knowledge about the system and modeling techniques, as well as to provide models that better approximate the actual brain.
Our model expands the toolset available for studying fruit flies, but the implications also extend beyond flies. Our work provides a foundation for using connectomes and machine learning to understand how biological neural networks process information efficiently. To accelerate this research and inspire experimentally testable predictions, we have made our tools available to the research community, enabling other researchers to use our model in their computational work.
Connectome-constrained models are compact artificial intelligences that are biologically plausible and map to their efficient biological counterparts. We’ve created a bridge between artificial and biological neural networks, opening new avenues for understanding both.
[1] Llama 3.1 70B’s training: 7.0 million H100-80GB hours at 700W per GPU: 7,000,000 hours × 0.7 kW = 4,900,000 kWh; An average U.S. household uses 1.22 kWh per hour: 4,900,000 kWh ÷ 1.22 kWh/household-hour ≈ 4,016,393 household-hours
Original publication:
Janne K. Lappalainen, Fabian D. Tschopp, Sridhama Prakhya, Mason McGill, Aljoscha Nern, Kazunori Shinomiya, Shinya Takemura, Eyal Gruntman, Jakob H. Macke, Srinivas C. Turaga. Connectome-constrained networks predict neural activity across the fly visual system. Nature 634, 1132–1140 (2024). https://doi.org/10.1038/s41586-024-07939-3
Cover illustration: Franz-Georg Stämmele
Human-guided Neural Networks for Synchrotron Experiments

Comments
Justin, December 21, 2024
Wonderful article Janne! A great set of guiding lessons-learned as we get better and better deep mechanistic models